The London Calling 2023 event featured three great talks about the use of nanopore sequencing to characterise a variety of infectious diseases in low-resource settings. Each of these presentations demonstrated how the portable MinION was being utilised to provide rapid sequencing capability to locations where, previously, samples may have had to be sent away for sequencing resulting in turnaround times of weeks.
This allows results to be generated at the sample source, providing real-time and detailed insights into the pathogen's genetic makeup, enabling rapid identification of its origin, transmission patterns, and potential mutations, thus guiding effective containment measures and public health response strategies.
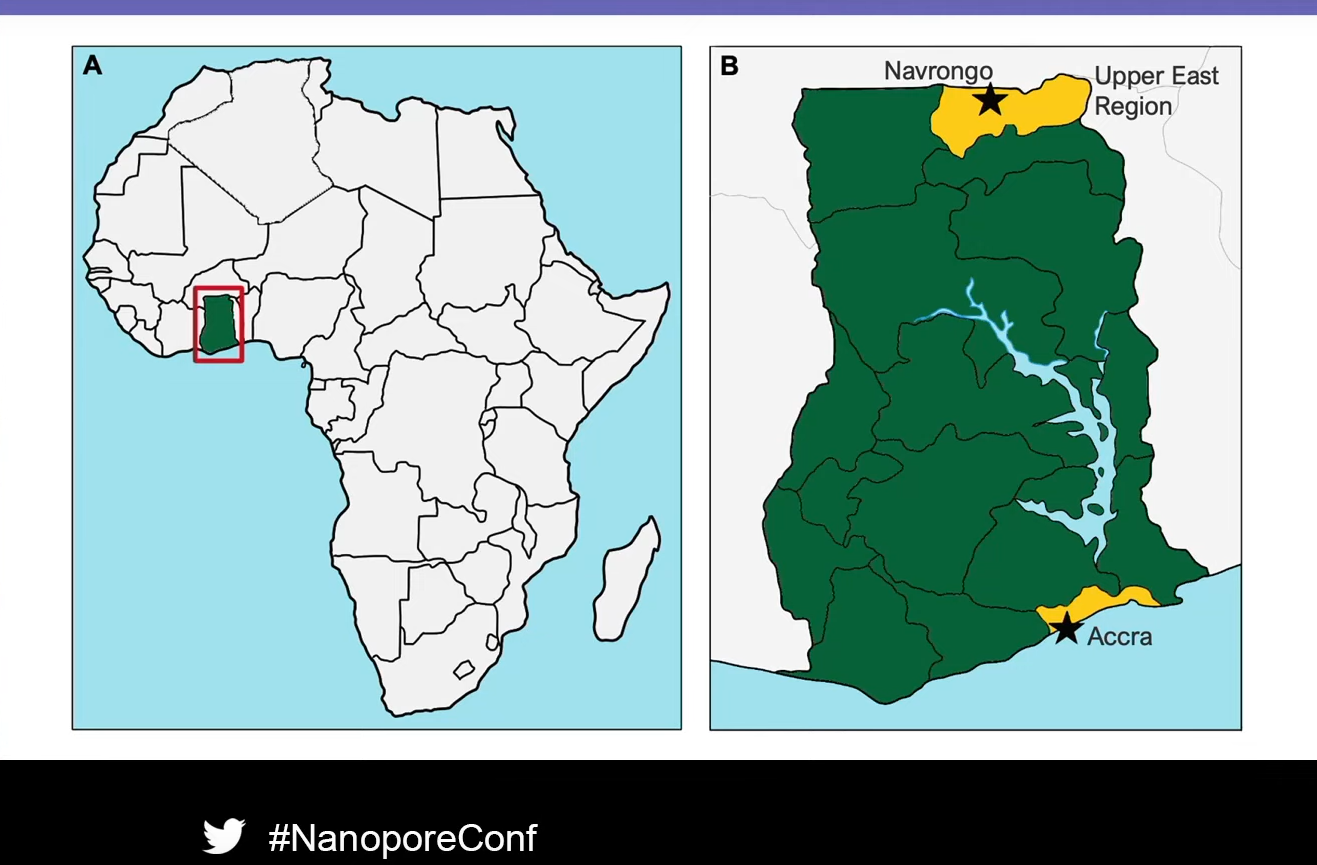
William Hamilton (Wellcome Sanger Institute, UK, and Cambridge University Department of Medicine, UK) delivers the talk highlighting results from the Nanopore sequencing for real-time genomic surveillance of Plasmodium falciparum project collaboratively led by himself and Dr Lucas Amenga-Etego (The West African Centre for Cell Biology of Infectious Pathogens (WACCBIP), University of Ghana) which leveraged on nanopore sequencing to explore resistance of malaria parasites to antimalarial drugs1.
Malaria, caused by Plasmodium falciparum, is a eukaryotic parasite that is transmitted to humans via Anopheles spp. mosquitoes, and can cause serious disease and death. William described how there were over 600,000 deaths attributed to malaria in 2021, with the greatest burden occurring in sub-Saharan Africa, where 95% of global malaria deaths occurred that year; with 76% of recorded deaths occurring in young children. There are drugs available for the treatment of malaria, but in some places, notably southeast Asia, the parasite has become resistant to multiple antimalarial drugs, including certain forms of the current front-line treatment, ACT (artemisinin-based combination therapy). Worryingly, parasites with partial resistance to artemisinins have been reported in East Africa in recent years. William commented that ‘The WHO has highlighted this as a major issue’ and emphasised the importance of surveillance to monitor the emergence and spread of resistance. However, checking for the presence of resistance in malaria parasites is difficult, as traditionally they need to be isolated from blood samples and tested in a laboratory — by contrast, genomics allows rapid access to resistance information without the need for lab culture. In addition to drug treatments, William explained that the first malaria vaccine (RTS,S) has recently been implemented in Ghana, Malawi, and Kenya, with plans for further rollout. The vaccine targets an antigen within the parasite called circumsporozoite protein (CSP); monitoring any changes in the diversity of the vaccine antigen target gene that occur as the vaccine is rolled out is one of the goals of the project. William explained that the further goals of the project were to perform nanopore sequencing in Ghana for malaria genomic surveillance, to describe the prevalence of drug resistance markers and to provide training and capacity-building for nanopore sequencing.
“Absolutely everything from sample collection, to processing, to sequencing, to bioinformatics analysis and final interpretation, all needed to be done in Ghana.... and we wanted to build a platform that was sustainable, that had a pretty simple and straightforward workflow that could be implemented in endemic settings”
The sequencing workflow consisted of DNA extraction followed by multiplex PCR and library preparation with the Native Barcoding Kit, and sequencing using a MinION with R10 MinION Flow Cells. Real-time basecalling was performed in super accuracy (SUP) mode with the Guppy basecaller and subsequent bioinformatics analysis involved the mapping of reads to reference sequences and variant calling with Medaka. Before testing the workflow at the sample source, the team validated their assay using mocked-up isolates with parasite gDNA spiked into human clinical research samples.
They found that even from low-volume blood research samples and at very low infection levels, the resistance genotype was correctly called with nanopore sequencing. William remarked that ‘we know what the genotype should be for all of these lab isolates because they are very well described, and we found complete agreement using R10 sequencing chemistry’. Although the data used in this study were from venous blood clinical research samples, William commented that they had tested the workflow on dried blood spots, which consist of small volumes of blood, and the assay worked very well.

The sequencing station at the Navrongo Health Research Centre
The workflow was then tested at two sites in Ghana: Accra, the capital city, and Navrongo, a rural town in the north. During the study period, the team collected 142 venous blood research samples and analysed 109 using the nanopore sequencing workflow. William went on to summarise the drug resistance results, remarking that they found very high coverage using their amplicon sequencing assay, with over 1000x coverage for each amplicon. Reassuringly, they found no parasites that were resistant to the current frontline antimalarial, artemisinin. However, there was a high frequency of resistance to sulfadoxine and pyrimethamine, which, William explained, are used together prophylactically to prevent malaria during pregnancy. Although ‘high-level’ resistance to the combination, which reduces the protective effect in pregnancy, was not found, the results highlight the need for ongoing surveillance. One particularly interesting result was the discovery that chloroquine resistance in malaria parasites, which was common in Ghana a decade ago, was not observed, and most parasites sequenced were chloroquine susceptible. William explained that this may be because chloroquine has not been used to treat malaria in Ghana for many years, so the resistance genotype has reverted.
Read the full project publication at
- Girgis, S. T., Adika, E., Nenyewodey, F. E., Senoo Jnr, D. K., Ngoi, J. M., Bandoh, K., ... & Hamilton, W. L. (2022). Nanopore sequencing for real-time genomic surveillance of Plasmodium falciparum. bioRxiv. DOI: https://doi.org/10.1101/2022.12.20.521122.
More talks from the London Calling 2023 event can read at https://nanoporetech.com/about-us/news/blog-nanopore-sequencing-disease-surveillance-low-resource-settings-updates-london